Translation Initiation Site Prediction
Biological Background
Biological Background
The main structural and functional molecules of an organism’s cell are proteins. Another family of molecules, nucleic acids, carry the genetic information. The most common nucleic acids are deoxyribonucleic acid (DNA) and ribonucleic acid (RNA). All these molecules are called macromolecules, due to their length. Both proteins and nucleic acids are linear polymers of smaller molecules (monomers). The term sequence is used to refer to the order of monomers that compose the macromolecule. A sequence can be represented as a string of different symbols, one for each monomer. There are twenty protein monomers called amino acids and five nucleic acid monomers called nucleotides (Table 1). DNA is the genetic material of almost every living organism. RNA has many functions inside a cell and plays an important role in protein synthesis. Moreover, RNA is the genetic material for some viruses. A sequence of nucleotides has two ends called the 5´ and the 3´ end. Moreover, it is directed from the 5´ to the 3´ end (5´® 3´).
Table 1. The five nucleotides are characterized by the nitrogenous base they contain. Three of them are present in both nucleic acids, one only in DNA and one only in RNA
|
Nitrogenous Base |
Symbol |
Nucleic Acid |
|
Adenine |
A |
DNA/RNA |
|
Cytosine |
C |
DNA/RNA |
|
Guanine |
G |
DNA/RNA |
|
Thymine |
T |
DNA |
|
Uracil |
U |
RNA |
The Central Dogma of
Molecular Biology
The central dogma of molecular biology
describes the flow of genetic information (Figure
1). DNA is transcribed into RNA and then RNA is
translated into proteins. The circular arrow around DNA denotes its
replication ability. However, RNA is reverse transcribed into DNA, in
retroviruses and RNA is able to replicate itself, in some viruses. These
cases are described by the dashed arrows. The replication of
nucleic acids is responsible for the transfer of genetic information from
parents to offspring. During transcription, DNA is used as template for
the synthesis of a messenger RNA (mRNA) molecule. This molecule carries
the initial “message” of DNA and under certain conditions is translated
into a protein molecule. In our setup, we focus on the process of
translation, which is further explained in the next paragraph.
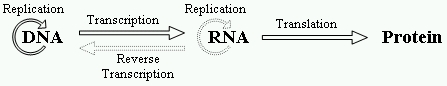
Figure 1.The central dogma of molecular biology
Translation
An organelle called ribosome, composed of ribosomal RNA (rRNA) and proteins,
is the “factory” where translation
takes place. The mRNA is scanned by the ribosome, which reads triplets of
nucleotides named codons. Thus, a
protein of N amino acids is coded by a sequence of 3N
nucleotides. Some amino acids are coded by more than one codons, because
there are 64 (43) different triplets for coding 20 amino
acids. Translation almost always initiates at an AUG codon (codes the
amino acid methionine) subsequent to the ribosome binding site. GUG and
UUG sometimes are used as start codons, but this rarely happens in
organisms other than bacteria [1]. Moreover, there are three stop codons
encoding the termination of translation (UAG, UAA and UGA). After the
initiation of translation, the ribosome “reads” the mRNA triplet by
triplet. A transfer RNA (tRNA) molecule brings the proper amino acid to
the protein synthesis site. The amino acid is added to the protein chain,
which, by this way, is elongated until a stop codon is reached. A protein
may go through post-translational modifications that affect its function.
Translation, usually, initiates at the AUG codon nearest to the 5´ end of the mRNA molecule. However, this does not happen in all cases. There are some escape mechanisms that allow the initiation of translation at following, but still near the 5´ end, AUG codons. One of them is leaky scanning, where the first AUG is bypassed due to inappropriate context. Another escape mechanism is reinitiation, where translation initiates at an AUG codon before the correct initiation site and ends by reaching a stop codon. Translation reinitiates when the true AUG codon is found. Sometimes direct internal initiation happens. In this case the ribosome directly attaches near the true AUG codon without any scanning. These mechanisms of the translation initiation process make more difficult the recognition of the TIS on a given genomic sequence.
There are three different ways to read a given sequence in a given direction. Each of these ways of reading is referred to as reading frame. The first reading frame starts at position 1, the second at position 2 and the third at position 3. The reading frame that is translated into a protein is named Open Reading Frame (ORF). A codon that is contained in the same reading frame with respect to another codon is referred to as “in-frame codon”. The coding region of an ORF is bounded by the initiation codon and the first in-frame stop codon. The coding region is surrounded by non-coding regions called 5´ and 3´ untranslated regions (UTRs). The direction of translation is 5´® 3´. We name upstream the region of a nucleotide sequence from a reference point towards the 5´ end. Respectively, the region of a nucleotide sequence from a reference point towards the 3´ end is referred to as downstream. For example, the initiation codon is upstream of the stop codon and the stop codon downstream of the initiation codon. In TIS classification problems the reference point is an AUG codon. The above are illustrated in Figure 2.
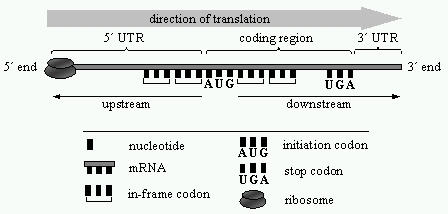
Figure 2. The initiation of translation. The ribosome scans the mRNA until it reads an AUG codon. If the AUG codon has appropriate context, then probably the translation initiates at that site
References
1. Kozak, M.: Initiation of Translation in Prokaryotes and Eukaryotes, Gene, 234(2), 187-208, 1999.